数据STUDIO转载|作者 刘秋言
特征筛选是建模过程中的重要一环。
基于决策树的算法,如Random Forest, Lightgbm, Xgboost,都能返回模型默认的Feature Importance,但诸多研究都表明该重要性是存在偏差的。
是否有更好的方法来筛选特征呢?Kaggle上很多大师级的选手通常采用的一个方法是Permutation Importance。这个想法最早是由Breiman (2001)[1]提出,后来由Fisher, Rudin, and Dominici (2018)改进[2]。
通过本文,你将通过一个Kaggle Amex真实数据了解到,模型默认的Feature Importance存在什么问题,什么是Permutation Importance,它的优劣势分别是什么,以及具体代码如何实现和使用。
另外,本文代码使用GPU来完成dataframe的处理,以及XGB模型的训练和预测。相较于CPU的版本,可以提升10到100倍的速度。具体来说,使用RAPIDS的CUDF来处理数据。训练模型使用XGB的GPU版本,同时使用DeviceQuantileDMatrix作为dataloader来减少内存的占用。做预测时,则使用RAPIDS的INF。
本文目录
模型默认的Feature Importance存在什么问题?
什么是Permutation Importance?
它的优劣势是什么?
Amex数据实例验证
01 模型默认的Feature Importance存在什么问题?
Strobl et al[3]在2007年就提出模型默认的Feature Importance会偏好连续型变量或高基数(high cardinality)的类型型变量。这也很好理解,因为连续型变量或高基数的类型变量在树节点上更容易找到一个切分点,换言之更容易过拟合。
另外一个问题是,Feature Importance的本质是训练好的模型对变量的依赖程度,它不代表变量在unseen data(比如测试集)上的泛化能力。特别当训练集和测试集的分布发生偏移时,模型默认的Feature Importance的偏差会更严重。
举一个极端的例子,如果我们随机生成一些X和二分类标签y,并用XGB不断迭代。随着迭代次数的增加,训练集的AUC将接近1,但是验证集上的AUC仍然会在0.5附近徘徊。这时模型默认的Feature Importance仍然会有一些变量的重要性特别高。这些变量帮助模型过拟合,从而在训练集上实现了接近1的AUC。但实际上这些变量还是无意义的。
02 什么是Permutation Importance?
Permutation Importance是一种变量筛选的方法。它有效地解决了上述提到的两个问题。
Permutation Importance将变量随机打乱来破坏变量和y原有的关系。如果打乱一个变量显著增加了模型在验证集上的loss,说明该变量很重要。如果打乱一个变量对模型在验证集上的loss没有影响,甚至还降低了loss,那么说明该变量对模型不重要,甚至是有害的。
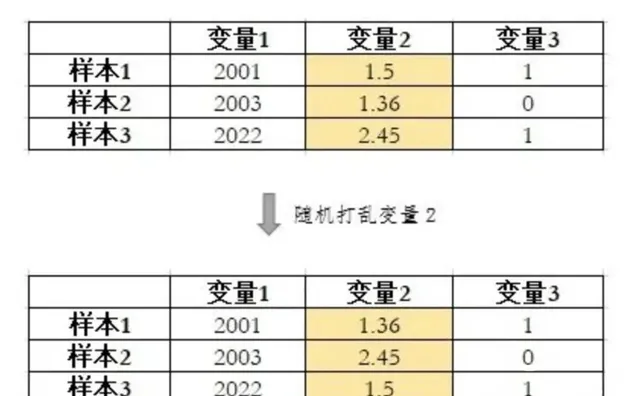
打乱变量示例
变量重要性的具体计算步骤如下:
1. 将数据分为train和validation两个数据集
2. 在train上训练模型,在validation上做预测,并评价模型(如计算AUC)
3. 循环计算每个变量的重要性:
(3.1) 在validation上对单个变量随机打乱;
(3.2)使用第2步训练好的模型,重新在validation做预测,并评价模型;
(3.3)计算第2步和第3.2步validation上模型评价的差异,得到该变量的重要性指标
Python代码步骤(model表示已经训练好的模型):
def permutation_importances(model, X, y, metric):
baseline = metric(model, X, y)
imp = []
for col in X.columns:
save = X[col].copy()
X[col] = np.random.permutation(X[col])
m = metric(model, X, y)
X[col] = save
imp.append(baseline - m)
return np.array(imp)
03 Permutation Importance的优劣势是什么?
3.1 优势
可以在任何模型上使用。不只是在基于决策树的模型,在线性回归,神经网络,任何模型上都可以使用。
不存在对连续型变量或高基数类别型变量的偏好。
体现了变量的泛化能力,当数据发生偏移时会特别有价值。
相较于循环的增加或剔除变量,不需要对模型重新训练,极大地降低了成本。但是循环地对模型做预测仍然会花费不少时间。
3.2 劣势
对变量的打乱存在随机性。这就要求随机打乱需要重复多次,以保证统计的显著性。
对相关性高的变量会低估重要性,模型默认的Feature Importance同样存在该问题。
04 A mex数据实例验证

理论听起来可能有点头痛,我们直接以Kaggle的Amex数据作为实例,验证下Permutation Importance的效果。为了方便阅读,代码附在文末!
考虑到Permutation Importance的随机性,我们将数据划分为10个fold,并且每个变量随机打乱10次,所以每个变量总共打乱100次,再计算打乱后模型评价差异的平均值,以保证统计上的显著性。之后我们可以再谈谈如何更好地随机打乱以节省资源。
这里我们分别对比3种变量重要性的排序:
模型默认的Feature Importance
Permutation Importance
标准化后的Permutation Importance:Permutation Importance / 随机100次的标准差。这考虑到了随机性。如果在Permutation Importance差不多的情况下,标准差更小,说明该变量的重要性结果是更稳定的。
为了简化实验,这里随机筛选了总共300个变量。使用300个变量时,模型10 fold AUC为0.9597。
下表是不同变化重要性排序下,模型AUC随着变量个数增加的变化情况:
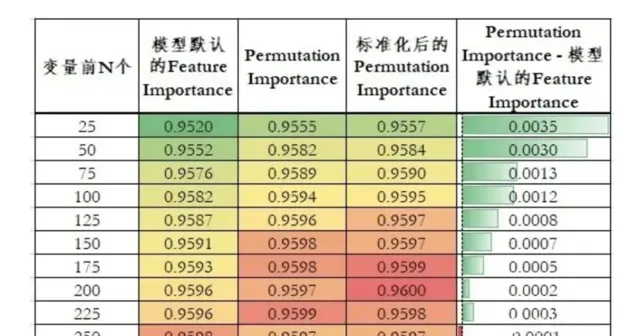
不同变量重要性排序下,模型效果的变化情况
由此我们可以得到如下结论:
Permutation Importance相较于模型默认的Feature Importance具有更好的排序性。当变量个数小于250个时,使用Permutation Importance排序的变量模型效果都更好。
随着变量个数的增加,模型默认的Feature Importance和Permutation Importance两种排序的模型AUC差异逐渐减小。这也间接说明Permutation Importance的重要性排序更好。因为在变量个数少的时候,两种排序筛选的变量差异会更大。随着变量的增加,两种排序下变量的重合逐渐增加,差异逐渐减小。
标准化后的Permutation Importance效果仅略微好于Permutation Importance,这在真实业务场景意义较低,在建模比赛中有一定价值。
代码附录
# !pip install kaggle
# !mkdir /root/.kaggle
# !cp /storage/kaggle.json /root/.kaggle/kaggle.json
# !chmod 600 /root/.kaggle/kaggle.json
# !kaggle datasets download -d raddar/amex-data-integer-dtypes-parquet-format -p /storage/data --unzip
import os
os.listdir('/storage/data')
['train.parquet',
'__notebook_source__.ipynb',
'cust_type.parquet',
'test.parquet',
'train_labels.csv']
import pandas as pd, numpy as np
pd.set_option('mode.chained_assignment', None)
import cupy, cudf
from numba import cuda
from cuml import ForestInference
import joblib
import time
import matplotlib.pyplot as plt
import pyarrow.parquet as pq
import random
from tqdm import tqdm
import pickle
import random
import gc
from sklearn.model_selection import train_test_split, KFold, StratifiedKFold
from sklearn.metrics import roc_auc_score
import xgboost as xgb
print('XGB Version',xgb.__version__)
XGB Version 1.6.1
CWD = '/storage/data'# 将CWD换成你的数据目录
SEED = 42
NAN_VALUE = -127# will fit in int8
FOLDS = 10
RAW_CAT_FEATURES = ["B_30","B_38","D_114","D_116","D_117","D_120","D_126","D_63","D_64","D_66","D_68"]
Utils
deffast_auc(y_true, y_prob):
y_true = cupy.asarray(y_true)
y_true = y_true[cupy.argsort(y_prob)]
cumfalses = cupy.cumsum(1-y_true)
nfalse = cumfalses[-1]
auc = (y_true * cumfalses).sum()
auc = auc / (nfalse * (len(y_true) - nfalse))
return auc
defeval_auc(preds, dtrain):
labels = dtrain.get_label()
return'auc', fast_auc(labels, preds), True
# NEEDED WITH DeviceQuantileDMatrix BELOW
classIterLoadForDMatrix(xgb.core.DataIter):
def__init__(self, df=None, features=None, target=None, batch_size=256*512):
self.features = features
self.target = target
self.df = df
self.it = 0# set iterator to 0
self.batch_size = batch_size
self.batches = int( np.ceil( len(df) / self.batch_size ) )
super().__init__()
defreset(self):
'''Reset the iterator'''
self.it = 0
defnext(self, input_data):
'''Yield next batch of data.'''
if self.it == self.batches:
return0# Return 0 when there's no more batch.
a = self.it * self.batch_size
b = min( (self.it + 1) * self.batch_size, len(self.df) )
dt = cudf.DataFrame(self.df.iloc[a:b])
input_data(data=dt[self.features], label=dt[self.target]) #, weight=dt['weight'])
self.it += 1
return1
deftrain_xgb_params(params, train, val_folds, features, early_stopping_rounds, num_boost_round, verbose_eval, data_seed, model_seed, ver='model', save_dir='./'):
importances = []
oof = []
skf = KFold(n_splits=FOLDS, shuffle=True, random_state=data_seed)
for fold in val_folds:
Xy_train = IterLoadForDMatrix(train.loc[train['fold'] != fold], features, 'target')
X_valid = train.loc[train['fold'] == fold, features]
y_valid = train.loc[train['fold'] == fold, 'target']
dtrain = xgb.DeviceQuantileDMatrix(Xy_train, max_bin=256)
dvalid = xgb.DMatrix(data=X_valid, label=y_valid)
# TRAIN MODEL FOLD K
# XGB MODEL PARAMETERS
xgb_parms = {
'eval_metric':'auc',
'objective':'binary:logistic',
'tree_method':'gpu_hist',
'predictor':'gpu_predictor',
'random_state':model_seed,
}
xgb_parms.update(params)
model = xgb.train(xgb_parms,
dtrain=dtrain,
evals=[(dtrain,'train'), (dvalid,'valid')],
num_boost_round=num_boost_round,
early_stopping_rounds=early_stopping_rounds,
verbose_eval=verbose_eval,
)
best_iter = model.best_iteration
if save_dir:
model.save_model(os.path.join(save_dir, f'{ver}_fold{fold}_{best_iter}.xgb'))
# GET FEATURE IMPORTANCE FOR FOLD K
dd = model.get_score(importance_type='weight')
df = pd.DataFrame({'feature':dd.keys(),f'importance_{fold}':dd.values()})
df = df.set_index('feature')
importances.append(df)
# INFER OOF FOLD K
oof_preds = model.predict(dvalid, iteration_range=(0,model.best_iteration+1))
if verbose_eval:
acc = fast_auc(y_valid.values, oof_preds)
print(f'acc_{fold}', acc)
# SAVE OOF
df = train.loc[train['fold'] == fold, ['customer_ID','target'] ].copy()
df['oof_pred'] = oof_preds
oof.append( df )
del Xy_train, df, X_valid, y_valid, dtrain, dvalid, model
_ = gc.collect()
importances = pd.concat(importances, axis=1)
oof = cudf.concat(oof, axis=0, ignore_index=True).set_index('customer_ID')
acc = fast_auc(oof.target.values, oof.oof_pred.values)
return acc, oof, importances
defclear_file(data_dir):
files = [c for c in os.listdir(data_dir) if'.ipynb_checkpoints'notin c]
for file in files:
os.remove(os.path.join(data_dir, file))
Split Data
train = cudf.read_csv(os.path.join(CWD, 'train_labels.csv'))
train['customer_ID'] = train['customer_ID'].str[-16:].str.hex_to_int().astype('int64')
train_cust = train[['customer_ID']]
CUST_SPLITS = train_cust.sample(frac=0.2, random_state=SEED)
CUST_SPLITS['fold'] = 9999
train_cv_cust = train_cust[~train_cust['customer_ID'].isin(CUST_SPLITS['customer_ID'].values)].reset_index(drop=True)
skf = KFold(n_splits=FOLDS, shuffle=True, random_state=SEED)
for fold,(train_idx, valid_idx) in enumerate(skf.split(train_cv_cust)):
df = train_cv_cust.loc[valid_idx, ['customer_ID']]
df['fold'] = fold
CUST_SPLITS = cudf.concat([CUST_SPLITS, df])
CUST_SPLITS = CUST_SPLITS.set_index('customer_ID')
CUST_SPLITS.to_csv('CUST_SPLITS.csv')
del train, train_cust
_ = gc.collect()
CUST_SPLITS['fold'].value_counts()
Data
defread_file(path = '', usecols = None):
# LOAD DATAFRAME
if usecols isnotNone: df = cudf.read_parquet(path, columns=usecols)
else: df = cudf.read_parquet(path)
# REDUCE DTYPE FOR CUSTOMER AND DATE
df['customer_ID'] = df['customer_ID'].str[-16:].str.hex_to_int().astype('int64')
df.S_2 = cudf.to_datetime( df.S_2 )
# SORT BY CUSTOMER AND DATE (so agg('last') works correctly)
#df = df.sort_values(['customer_ID','S_2'])
#df = df.reset_index(drop=True)
# FILL NAN
df = df.fillna(NAN_VALUE)
print('shape of data:', df.shape)
return df
print('Reading train data...')
train = read_file(path=os.path.join(CWD, 'train.parquet'))
defprocess_and_feature_engineer(df):
# FEATURE ENGINEERING FROM
# https://www.kaggle.com/code/huseyincot/amex-agg-data-how-it-created
all_cols = [c for c in list(df.columns) if c notin ['customer_ID','S_2']]
cat_features = ["B_30","B_38","D_114","D_116","D_117","D_120","D_126","D_63","D_64","D_66","D_68"]
num_features = [col for col in all_cols if col notin cat_features]
test_num_agg = df.groupby("customer_ID")[num_features].agg(['mean', 'std', 'min', 'max', 'last'])
test_num_agg.columns = ['_'.join(x) for x in test_num_agg.columns]
test_cat_agg = df.groupby("customer_ID")[cat_features].agg(['count', 'last', 'nunique'])
test_cat_agg.columns = ['_'.join(x) for x in test_cat_agg.columns]
df = cudf.concat([test_num_agg, test_cat_agg], axis=1)
del test_num_agg, test_cat_agg
print('shape after engineering', df.shape )
return df
train = process_and_feature_engineer(train)
train = train.fillna(NAN_VALUE)
# ADD TARGETS
targets = cudf.read_csv(os.path.join(CWD, 'train_labels.csv'))
targets['customer_ID'] = targets['customer_ID'].str[-16:].str.hex_to_int().astype('int64')
targets = targets.set_index('customer_ID')
train = train.merge(targets, left_index=True, right_index=True, how='left')
train.target = train.target.astype('int8')
del targets
# NEEDED TO MAKE CV DETERMINISTIC (cudf merge above randomly shuffles rows)
train = train.sort_index().reset_index()
# FEATURES
FEATURES = list(train.columns[1:-1])
print(f'There are {len(FEATURES)} features!')
Reading train data...
shape of data: (5531451, 190)
shape after engineering (458913, 918)
There are 918 features!
# ADD CUST SPLITS
CUST_SPLITS = cudf.read_csv('CUST_SPLITS.csv').set_index('customer_ID')
train = train.set_index('customer_ID').merge(CUST_SPLITS, left_index=True, right_index=True, how='left')
test = train[train['fold'] == 9999]
train = train[train['fold'] != 9999]
train = train.sort_index().reset_index()
train.head()
Permutation
defget_permutation_importance(model_path, data, model_features, shuffle_features, shuffle_times=5):
permutation_importance = pd.DataFrame(columns=['feature', 'metric', 'shuffle_idx'])
model = ForestInference.load(model_path, model_type='xgboost', output_ class=True)
preds = model.predict_proba(data[model_features])[1].values
acc = fast_auc(data['target'].values, preds)
permutation_importance.loc[permutation_importance.shape[0]] = ['original', cupy.asnumpy(acc), 0]
for col in tqdm(shuffle_features):
value = data[col].copy().values
for i in np.arange(shuffle_times):
np.random.seed(i)
data[col] = cupy.random.permutation(data[col].copy().values)
preds = model.predict_proba(data[model_features])[1].values
new_acc = fast_auc(data['target'].values, preds)
permutation_importance.loc[permutation_importance.shape[0]] = [col, cupy.asnumpy(new_acc), i]
data[col] = value
return permutation_importance
defagg_permu_files(permu_dir, count_threshold, retrain_idx):
permu_files = [c for c in os.listdir(permu_dir) if ('.csv'in c) and (f'permutation_importance_{retrain_idx}'in c)]
permutation_importance_all = []
for file in permu_files:
df = pd.read_csv(os.path.join(permu_dir, file))
df['permut_idx'] = file.split('_')[-2]
permutation_importance_all.append(df)
permutation_importance_all = pd.concat(permutation_importance_all)
# original_acc
original_acc = permutation_importance_all[permutation_importance_all['feature'] == 'original'].set_index(['fold', 'permut_idx'])[['metric']]
original_acc.rename({'metric': 'metric_ori'}, axis=1, inplace=True)
permutation_importance_all = permutation_importance_all.set_index(['fold', 'permut_idx']).merge(original_acc, left_index=True, right_index=True, how='left')
permutation_importance_all['metric_diff_ratio'] = (permutation_importance_all['metric_ori'] - permutation_importance_all['metric']) / permutation_importance_all['metric_ori']
permutation_importance_all.reset_index(inplace=True)
# random_acc
random_acc = permutation_importance_all[permutation_importance_all['feature'] == 'random'].groupby(['permut_idx'])[['metric_diff_ratio']].agg(['mean', 'std'])
random_acc.columns = ['random_mean', 'random_std']
permutation_importance_all.reset_index(inplace=True)
permutation_importance_agg = permutation_importance_all[permutation_importance_all['feature'] != 'random'].groupby(['feature', 'permut_idx'])['metric_diff_ratio'].agg(['min', 'max','mean', 'std', 'count'])
permutation_importance_agg['z'] = permutation_importance_agg['mean'] / permutation_importance_agg['std']
if count_threshold:
permutation_importance_agg = permutation_importance_agg[permutation_importance_agg['count'] == count_threshold]
permutation_importance_agg = permutation_importance_agg.reset_index().set_index('permut_idx')
permutation_importance_agg['z'] = permutation_importance_agg['mean'] / permutation_importance_agg['std']
permutation_importance_agg = permutation_importance_agg.merge(random_acc, left_index=True, right_index=True, how='left')
permutation_importance_agg['random_z'] = permutation_importance_agg['random_mean'] / permutation_importance_agg['random_std']
permutation_importance_agg = permutation_importance_agg.reset_index().set_index('feature')
return permutation_importance_agg
params_13m = {
'max_depth':4,
'learning_rate':0.1,
'subsample':0.8,
'colsample_bytree':0.1,
}
# ADD RANDOM FEATURE
np.random.seed(SEED)
train['random'] = np.random.normal(loc=1, scale=1, size=train.shape[0])
np.random.seed(SEED)
np.random.shuffle(FEATURES)
FEATURES = FEATURES[:300]
print(FEATURES[:10])
['D_107_max', 'D_41_mean', 'D_77_min', 'R_19_mean', 'D_131_min', 'B_41_max', 'D_76_max', 'D_91_max', 'D_54_mean', 'D_84_max']
permu_dir = './permut_13m_ver001_retrain'
ifnot os.path.isdir(permu_dir): os.mkdir(permu_dir)
clear_file(permu_dir)
retrain_idx = 1
val_folds = [0,1,2,3,4,5,6,7,8,9]
shuffle_times = 10
whileTrue:
# Features
sub_features_num = len(FEATURES)
feature_split_num = max(round(len(FEATURES) / sub_features_num), 1)
sub_features_num = len(FEATURES) // feature_split_num
print(f'FEATURES: {len(FEATURES)}; retrain_idx: {retrain_idx}; feature_split_num: {feature_split_num}; sub_features_num: {sub_features_num}')
start_idx = 0
for sub_features_idx in range(feature_split_num):
if sub_features_idx == feature_split_num - 1:
end_idx = len(FEATURES)
else:
end_idx = start_idx+sub_features_num
sub_features = FEATURES[start_idx: end_idx]
# Train
acc, oof, importances = train_xgb_params(params_13m,
train,
features=sub_features+['random'],
val_folds=val_folds,
early_stopping_rounds=100,
num_boost_round=9999,
verbose_eval=False,
data_seed=SEED,
model_seed=SEED,
save_dir=permu_dir,
ver=f'13M_{retrain_idx}_{sub_features_idx}')
importances.to_csv(os.path.join(permu_dir, 'original_importances.csv'))
# Permutation Importance
permutation_importance_list = []
for fold in val_folds:
print(f'........ sub_features_idx: {sub_features_idx}; start_idx: {start_idx}, end_idx: {end_idx}, fold: {fold};')
model_file = [c for c in os.listdir(permu_dir) iff'13M_{retrain_idx}_{sub_features_idx}_fold{fold}'in c]
if len(model_file) > 1:
raise ValueError(f'There are more than one model file: {model_file}')
model_file = model_file[0]
model = xgb.Booster()
model_path = os.path.join(permu_dir, model_file)
# PERMUTATION IMPORTANCE
importances_fold = importances[f'importance_{fold}']
shuffle_features = importances_fold[importances_fold.notnull()].index.to_list()
permutation_importance = get_permutation_importance(model_path,
train.loc[train['fold'] == fold, :],
model_features=sub_features+['random'],
shuffle_features=shuffle_features,
shuffle_times=shuffle_times)
permutation_importance['retrain_idx'] = retrain_idx
permutation_importance['sub_features_idx'] = sub_features_idx
permutation_importance['fold'] = fold
permutation_importance.to_csv(os.path.join(permu_dir, f'permutation_importance_{retrain_idx}_{sub_features_idx}_fold{fold}.csv'), index=False)
permutation_importance_list.append(permutation_importance)
start_idx = end_idx
permutation_importance_agg = agg_permu_files(permu_dir, count_threshold=len(val_folds)*shuffle_times, retrain_idx=retrain_idx)
drop_features = permutation_importance_agg[permutation_importance_agg['z'] < permutation_importance_agg['random_z']].index.to_list()
permutation_importance_agg['is_drop'] = permutation_importance_agg.index.isin(drop_features)
permutation_importance_agg.to_csv(os.path.join(permu_dir, f'permutation_importance_agg_{retrain_idx}.csv'))
FEATURES = [c for c in FEATURES if c notin drop_features]
retrain_idx += 1
break
# if len(drop_features) == 0:
# break
Baseline
val_folds = [0,1,2,3,4,5,6,7,8,9]
acc, oof, importances = train_xgb_params(params_13m,
train,
features=FEATURES,
val_folds=val_folds,
early_stopping_rounds=100,
num_boost_round=9999,
verbose_eval=False,
data_seed=SEED,
model_seed=SEED,
save_dir=False)
print('num_features', len(FEATURES), 'auc', acc)
num_features 300 auc 0.9597397304150984
筛选变量实验
val_folds = [0,1,2,3,4,5,6,7,8,9]
默认的Feature Importance
original_importances = pd.read_csv(os.path.join(permu_dir, 'original_importances.csv'))
original_importances = original_importances.set_index('feature')
original_importances['mean'] = original_importances.mean(axis=1)
original_importances['std'] = original_importances.std(axis=1)
original_importances.reset_index(inplace=True)
features_mean = original_importances.sort_values('mean', ascending=False)['feature'].to_list()
num_features = 25
whileTrue:
acc, oof, importances = train_xgb_params(params_13m,
train,
features=features_mean[:num_features],
val_folds=val_folds,
early_stopping_rounds=100,
num_boost_round=9999,
verbose_eval=False,
data_seed=SEED,
model_seed=SEED,
save_dir=False)
print('num_features', num_features, 'auc', acc)
num_features += 25
if num_features > permutation_importance_agg.shape[0]:
break
num_features 25 auc 0.9519786413209202
num_features 50 auc 0.9551949305682249
num_features 75 auc 0.957583263622448
num_features 100 auc 0.9581601072672994
num_features 125 auc 0.958722652426215
num_features 150 auc 0.959060598689679
num_features 175 auc 0.9593140320399974
num_features 200 auc 0.9595746364492562
num_features 225 auc 0.9596106789864496
num_features 250 auc 0.959798236138387
平均下降比例
permutation_importance_agg = pd.read_csv(os.path.join(permu_dir, 'permutation_importance_agg_1.csv'))
features_mean = permutation_importance_agg.sort_values('mean', ascending=False)['feature'].to_list()
num_features = 25
whileTrue:
acc, oof, importances = train_xgb_params(params_13m,
train,
features=features_mean[:num_features],
val_folds=val_folds,
early_stopping_rounds=100,
num_boost_round=9999,
verbose_eval=False,
data_seed=SEED,
model_seed=SEED,
save_dir=False)
print('num_features', num_features, 'auc', acc)
num_features += 25
if num_features > permutation_importance_agg.shape[0]:
break
num_features 25 auc 0.955488431492996
num_features 50 auc 0.9582052598376949
num_features 75 auc 0.9589016974455882
num_features 100 auc 0.9593989000375628
num_features 125 auc 0.9595675126936822
num_features 150 auc 0.9597684801181003
num_features 175 auc 0.9598145119359532
num_features 200 auc 0.9597407810213192
num_features 225 auc 0.9598664237091002
num_features 250 auc 0.959691966613858
标准化后的平均下降比例
permutation_importance_agg = pd.read_csv(os.path.join(permu_dir, 'permutation_importance_agg_1.csv'))
features_z = permutation_importance_agg.sort_values('z', ascending=False)['feature'].to_list()
num_features = 25
whileTrue:
acc, oof, importances = train_xgb_params(params_13m,
train,
features=features_z[:num_features],
val_folds=val_folds,
early_stopping_rounds=100,
num_boost_round=9999,
verbose_eval=False,
data_seed=SEED,
model_seed=SEED,
save_dir=False)
print('num_features', num_features, 'auc', acc)
num_features += 25
if num_features > permutation_importance_agg.shape[0]:
break
num_features 25 auc 0.9556982881629269
num_features 50 auc 0.9584120117990971
num_features 75 auc 0.9589902318791576
num_features 100 auc 0.9595072118772046
num_features 125 auc 0.9597150474489736
num_features 150 auc 0.9597403596967278
num_features 175 auc 0.9599117083347268
num_features 200 auc 0.9599500114626974
num_features 225 auc 0.9598069184316719
num_features 250 auc 0.959729845054988
参考
1. Breiman, Leo.「Random Forests.」 Machine Learning 45 (1). Springer: 5-32 (2001).
2. Fisher, Aaron, Cynthia Rudin, and Francesca Dominici. 「All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously.」 http://arxiv.org/abs/1801.01489 (2018).
3. S trobl C, et al. B ias in random forest variable importance measures: Illustrations, sources and a solution, BMC Bioinformatics, 2007, vol. 8 pg. 25
来源:https://zhuanlan.zhihu.com/p/563422607
--end--











